
Data-driven variable selection
Source:vignettes/articles/variable-selection.Rmd
variable-selection.RmdIntro
In the previous articles you have learned the core functions of SDMtune for training, evaluating and display the output of a model. In this article you will learn how to perform data-driven variable selection.
Explore variable correlation
We extract 10000 background locations using the function randomPoints
included in the dismo package (we set the seed to have
reproducible results). After we create an SWD() object
using the prepareSWD() function:
set.seed(25)
bg <- terra::spatSample(predictors,
size = 10000,
method = "random",
na.rm = TRUE,
xy = TRUE,
values = FALSE)
bg <- prepareSWD(species = "Bgs",
a = bg,
env = predictors,
categorical = "biome")#> Warning: [spatSample] fewer cells returned than requestedThe environmental variables we downloaded have a coarse resolution and the function can extract a bit less than 10000 random locations (see the warning message).
With the function plotCor() you can create an heat map
showing the degree of autocorrelation:
plotCor(bg,
method = "spearman",
cor_th = 0.7)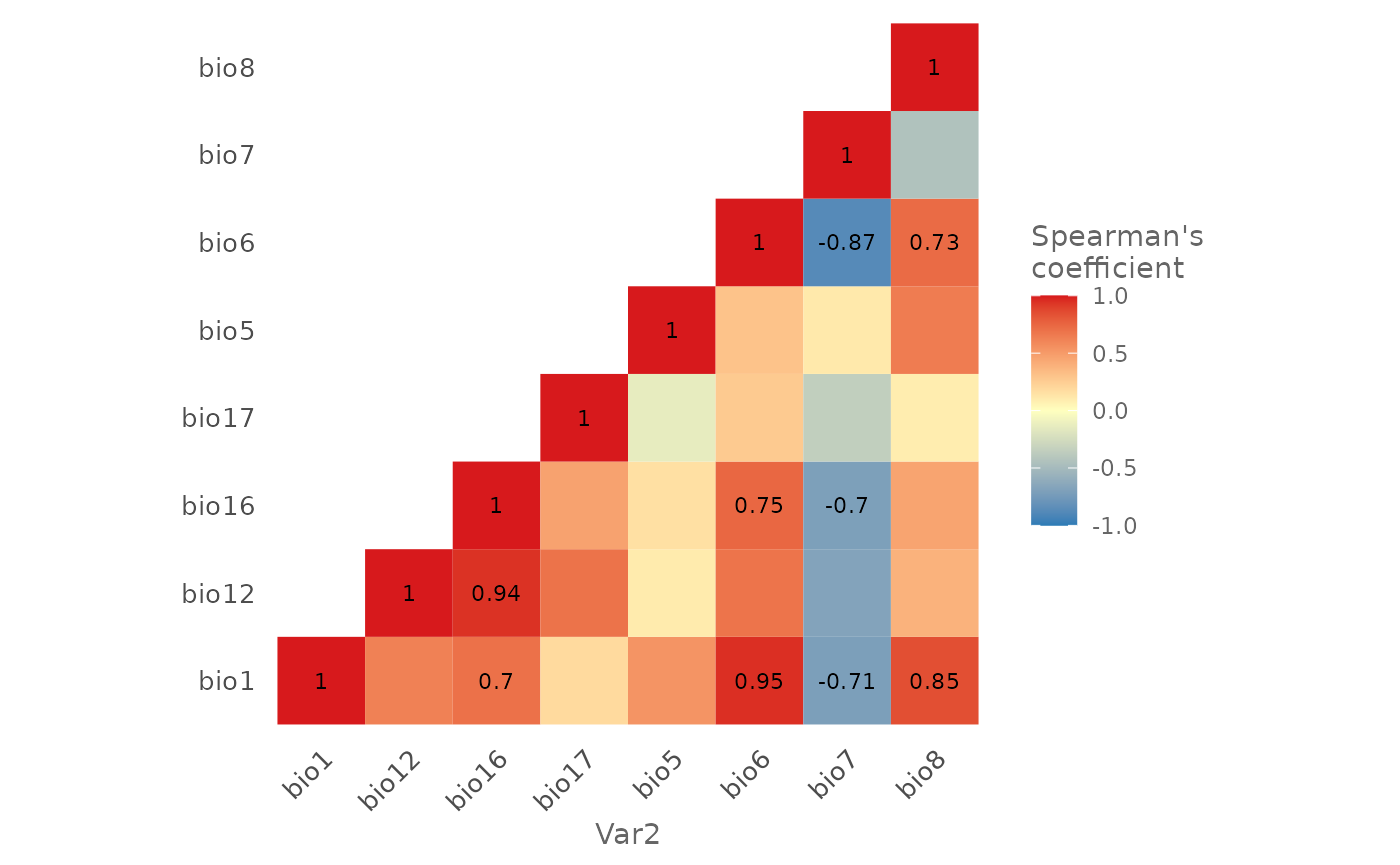
You can select a different correlation method or set a different
correlation threshold. Another useful function is corVar()
that instead of creating a heat map prints the pairs of correlated
variables according to the given method and correlation threshold:
corVar(bg,
method = "spearman",
cor_th = 0.7)| Var1 | Var2 | value |
|---|---|---|
| bio1 | bio6 | 0.9513541 |
| bio12 | bio16 | 0.9447559 |
| bio6 | bio7 | -0.8734498 |
| bio1 | bio8 | 0.8459649 |
| bio16 | bio6 | 0.7471269 |
| bio6 | bio8 | 0.7286723 |
| bio1 | bio7 | -0.7119135 |
| bio16 | bio7 | -0.7027568 |
| bio1 | bio16 | 0.7023585 |
As you can see there are few variables that have a correlation coefficient greater than 0.7 in absolute value.
Remove highly correlated variables
SDMtune implements an algorithm that removes highly correlated variables repeating the following steps:
- ranks the variables according to the permutation importance or the percent contribution (the second method is available only for Maxent models).
- checks if the variable ranked as most important is highly correlated with other variables, according to the given method and correlation threshold. If the algorithm finds correlated variables it moves to the next step, otherwise checks the other variables in the rank;
- performs a leave one out Jackknife test among the correlated variables;
- remove the variable that decreases the less the model performance when removed, according to the given metric on the training dataset.
The process is repeated until the remaining variables have a correlation coefficient lower than the provided correlation threshold. In the next example we remove the variables that have a Spearman correlation coefficient and checking the AUC on the training dataset (we use only one permutation to save time but you are free to increase this value). If you use RStudio, the function creates an interactive real-time chart in the viewer pane. Run the following code and hover over the chart during the execution of the function to get extra information:
selected_variables_model <- varSel(maxnet_model,
metric = "auc",
test = test,
bg4cor = bg,
method = "spearman",
cor_th = 0.7,
permut = 1)#> ✔ The variables bio16, bio6, bio7, and bio8 have been removedAs you can see some variables have been removed. The output of the function is the model trained with the selected variables:
selected_variables_model
#>
#> ── Object of class: <SDMmodel> ──
#>
#> Method: Maxnet
#>
#> ── Hyperparameters
#> • fc: "lqph"
#> • reg: 1
#>
#> ── Info
#> • Species: Virtual species
#> • Presence locations: 320
#> • Absence locations: 5000
#>
#> ── Variables
#> • Continuous: "bio1", "bio12", "bio17", and "bio5"
#> • Categorical: "biome"Try yourself
Remove highly correlated variables using the
default_model, ranking the variables with the percent
contribution and using the AICc as evaluation metric. Check the help of
the function varSel() and highlight the next cell to see
the solution:
# You need to pass the env argument for the AICc and the use_pc argument to use the percent contribution
selected_variables_model <- varSel(default_model,
metric = "aicc",
bg4cor = bg,
method = "spearman",
cor_th = 0.7,
env = predictors,
use_pc = TRUE)Remove variables with low importance
There are cases in which a model has some environmental variables
ranked with very low contribution and you may want to remove some of
them to reduce the model complexity. SDMtune offers two
different strategies implemented in the function
reduceVar(). We will use the maxnet_model
trained with all the variables. Let’s first check the permutation
importance (we use only one permutation to save time):
varImp(maxnet_model,
permut = 1)| Variable | Permutation_importance |
|---|---|
| bio1 | 56.1 |
| bio8 | 19.5 |
| bio17 | 6.3 |
| biome | 4.7 |
| bio12 | 3.5 |
| bio5 | 3.5 |
| bio7 | 3.2 |
| bio6 | 1.6 |
| bio16 | 1.5 |
We will use the function reduceVar() only for
demonstration purpose. In the first example we want to remove all the
environmental variables that have a permutation importance lower than
6%, no matter if the model performance decreases. The function removes
the last ranked environmental variable, trains a new model and computes
a new rank. The process is repeated until all the remaining
environmental variables have an importance greater than 6%:
cat("Testing AUC before: ", auc(maxnet_model, test = test))
#> Testing AUC before: 0.8505888
reduced_variables_model <- reduceVar(maxnet_model,
th = 6,
metric = "auc",
test = test,
permut = 1)
#> ✔ The variables bio16, bio6, bio7, bio5, bio17, and biome have been removed
cat("Testing AUC after: ", auc(reduced_variables_model, test = test))
#> Testing AUC after: 0.8520787In the second example we want to remove the environmental variables that have a permutation importance lower than 15% only if removing the variables the model performance does not decrease, according to the given metric. In this case the function performs a leave one out Jackknife test and remove the environmental variables in a step-wise fashion as described in the previous example, but only if the model performance doesn’t drop:
cat("Testing AUC before: ", auc(maxnet_model, test = test))
#> Testing AUC before: 0.8505888
reduced_variables_model <- reduceVar(maxnet_model,
th = 15,
metric = "auc",
test = test,
permut = 1,
use_jk = TRUE)
#> ✔ The variables bio16, bio6, bio5, bio12, biome, and bio8 have been removed
cat("Testing AUC after: ", auc(reduced_variables_model, test = test))
#> Testing AUC after: 0.8533188As you can see in this case several variables have been removed and the AUC in the testing dataset didn’t decrease.
Try yourself
Reduce the number of variables using the model trained with the cross validation and using as metric the TSS. Highlight the following cell for the solution:
# You need to pass TRUE to the test argument
selected_variables_model <- reduceVar(cv_model,
th = 6,
metric = "tss",
test = TRUE,
permut = 1)Conclusion
In this article you have learned:
- how to explore the correlation between the environmental variables;
- a possible approach to remove highly correlated environmental variables;
- two possible methods to remove environmental variables with low importance.
In the next article you will learn how to tune the model hyperparameters.